Methods Used to Study Trx-h
About Trx-h
- Originally found in cytosol
- Also found (to less extent) at other subcellular locations such as in the mitochondrium, ER, Golgi apparatus, and nucleus
- The largest TRX gene family in plants; 9 isoforms in A. thaliana
- Reduced by NTRA/NTRB
- NADPH produced by the oxidative pentose phosphate pathway (OPPP)
Trx-h targets
- Storage
- Redox control
- Amino acid and carbohydrate metabolism
- Proteinaceous inhibitors of α-amylases and proteases
- ...
mBBr Labelling
mBBr Labelling
- Fluorescent alkylating agent monobromobimane
- Label free -SH groups (in proteins reduced by Trx)
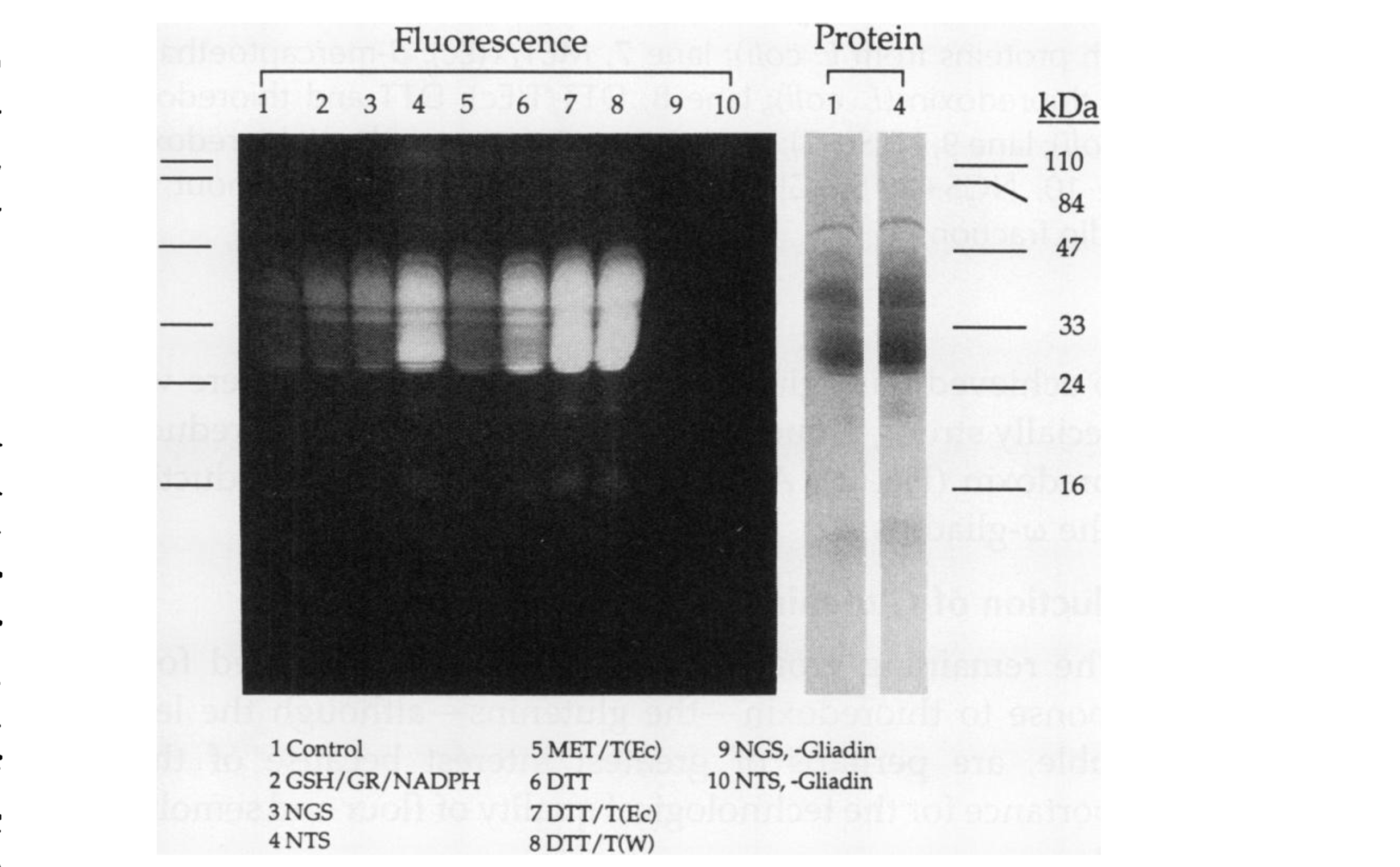
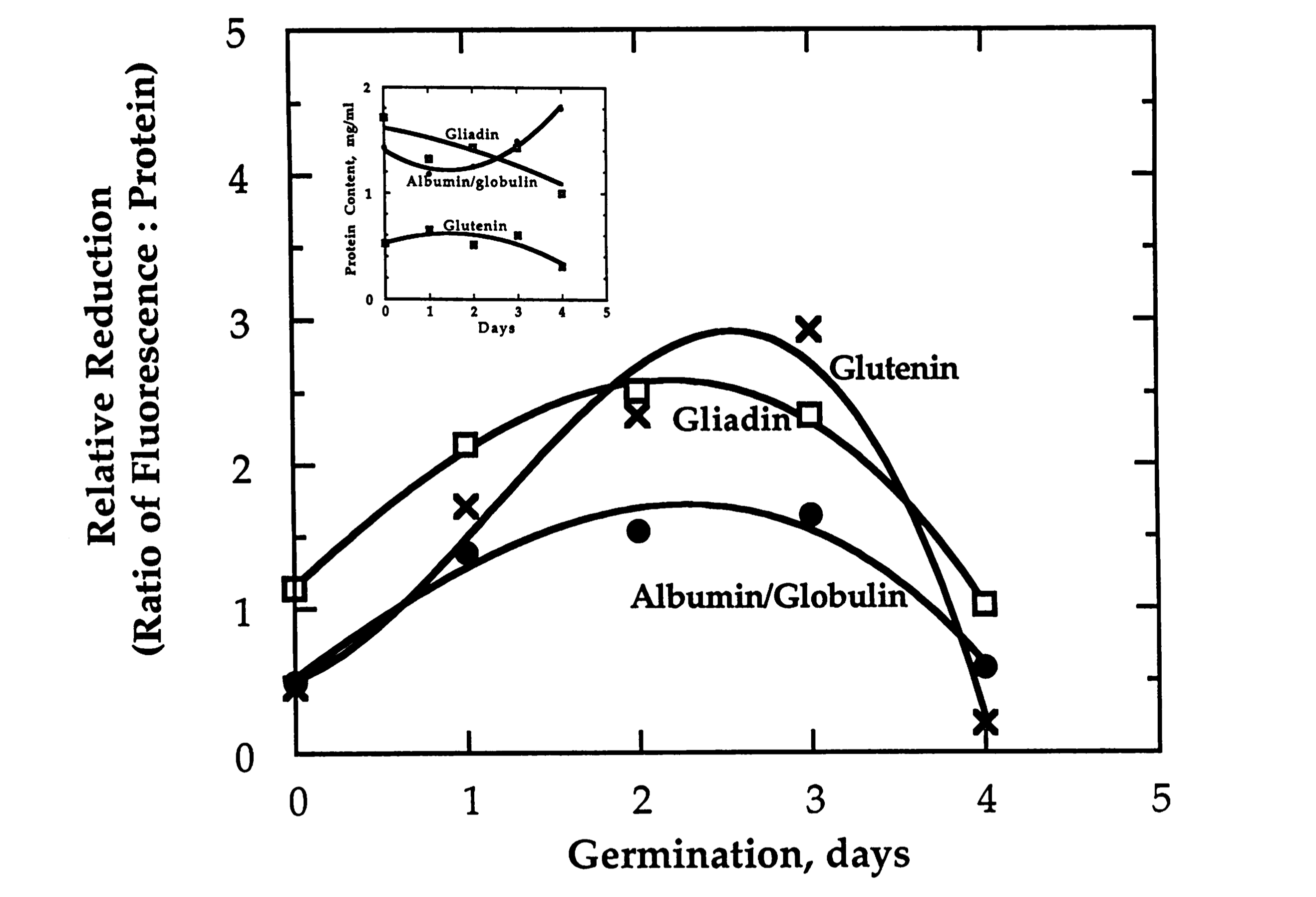
- Example: Specific Reduction of Wheat Storage Thioredoxin h1 (Kobrehel-1992)
CXXS Trx mutants
CXXS Trx mutants
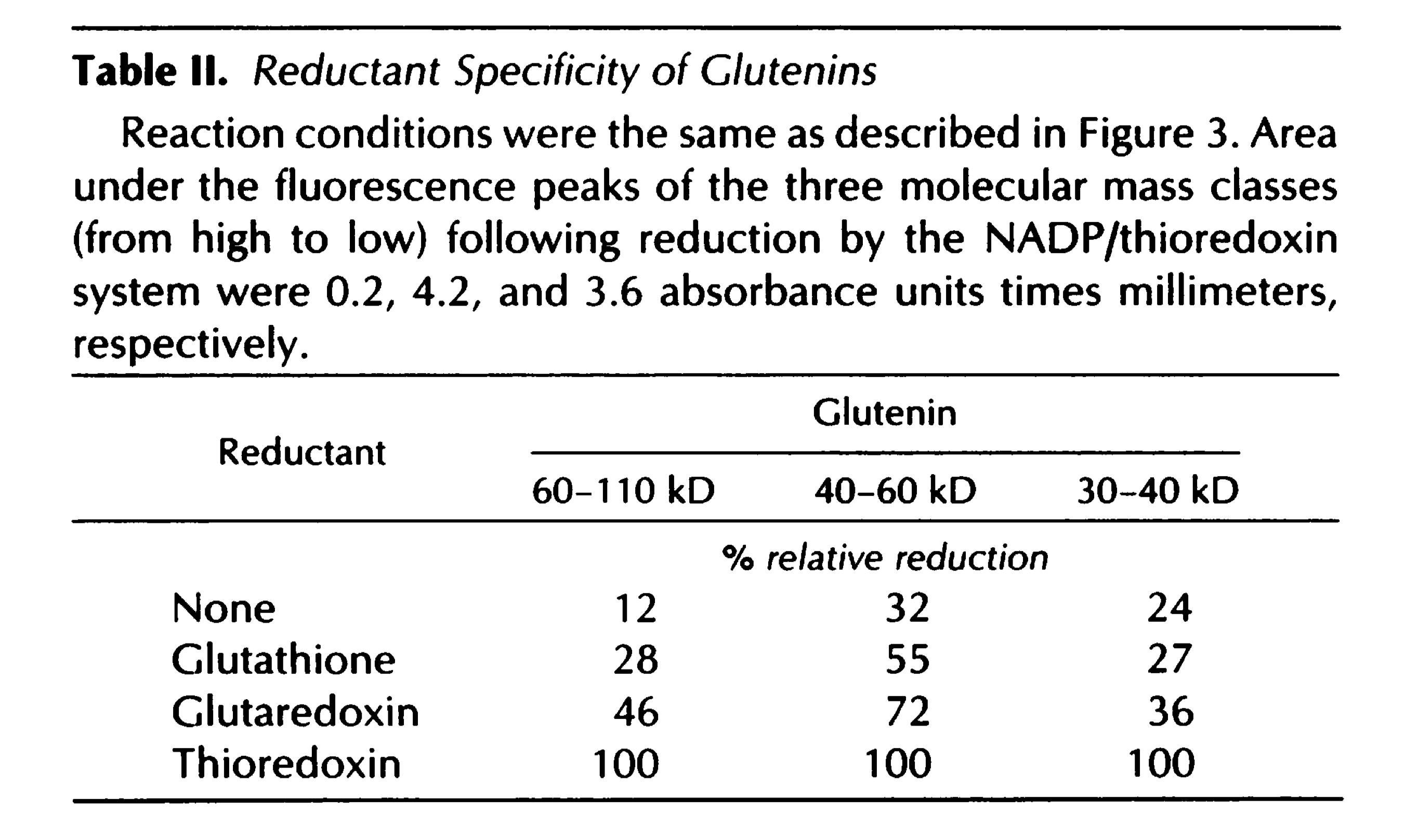
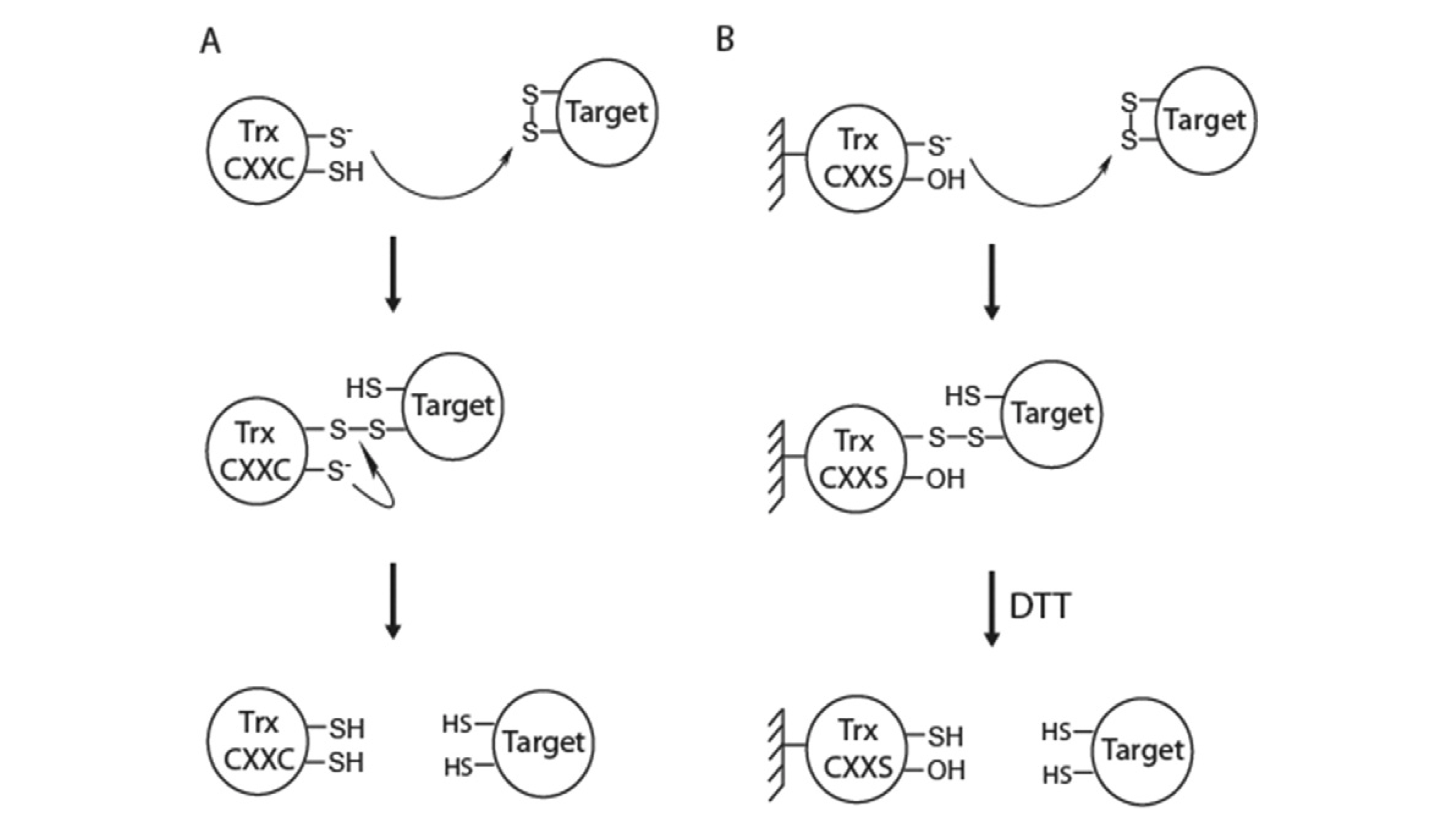
- Reduction of the target disulfide bond by Trx has two steps
- If one of the C in CXXC is replaced by S, the intermediate is trapped
- Originally applied by Verdoucq (1999)
- Improved by Yamazaki (2004): Immobilised (on Sepharose 4B) monocysteinic Trx-h traps targets in column; trapped targets subsequently eluted with DTT
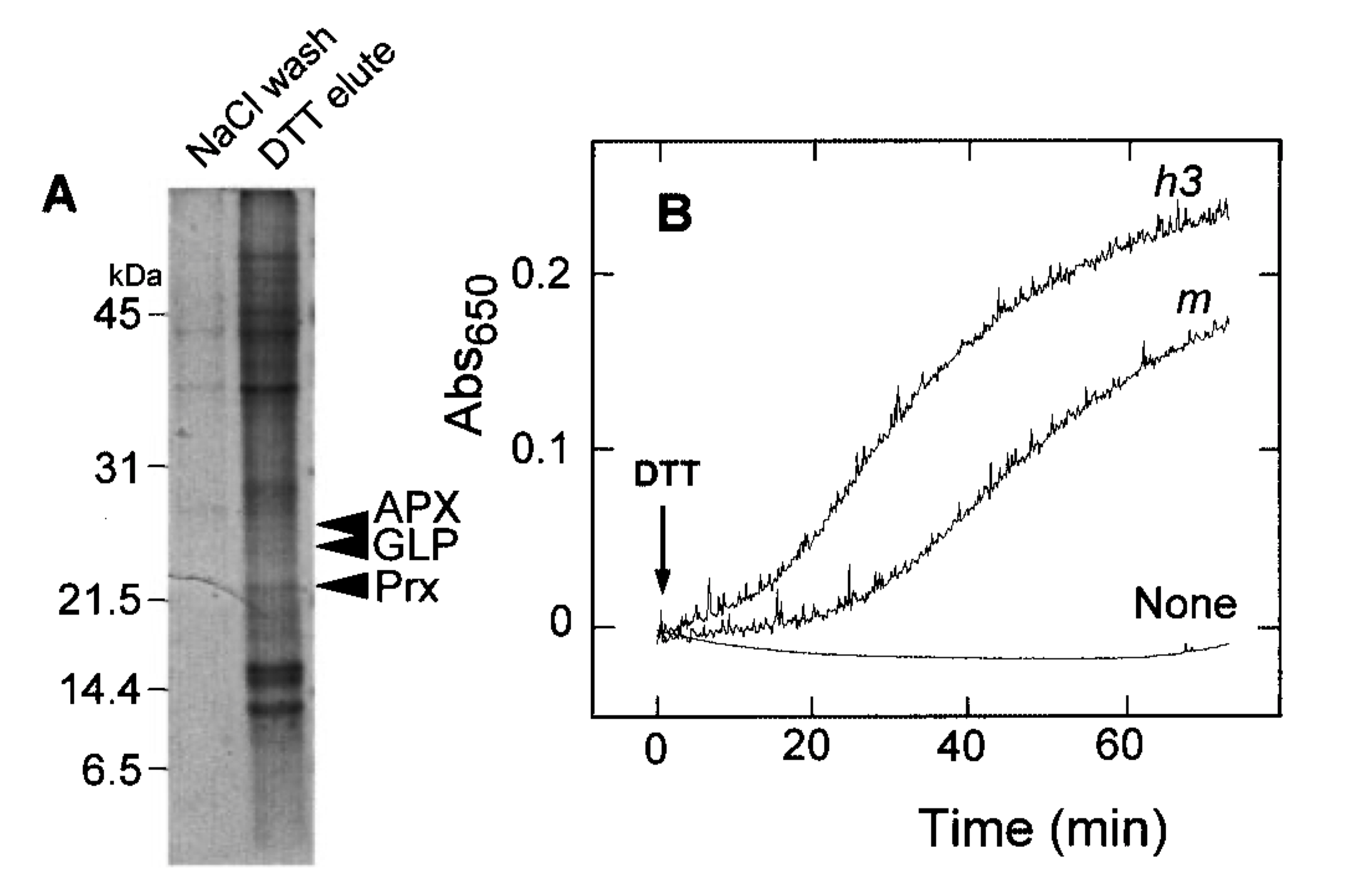
Differential Labelling
Determination of Target Disulphide Bonds
- Studies on chloroplastic Trxs mainly depended on site directed mutagenesis
- Trx h has too many targets; mutagenesis would be inefficient
- MS can find positions of chemical modifications
- Normal MS cannot distinguish between -SH and -S-S-
- ...because of complete reduction and alkylation prior to sample preparation
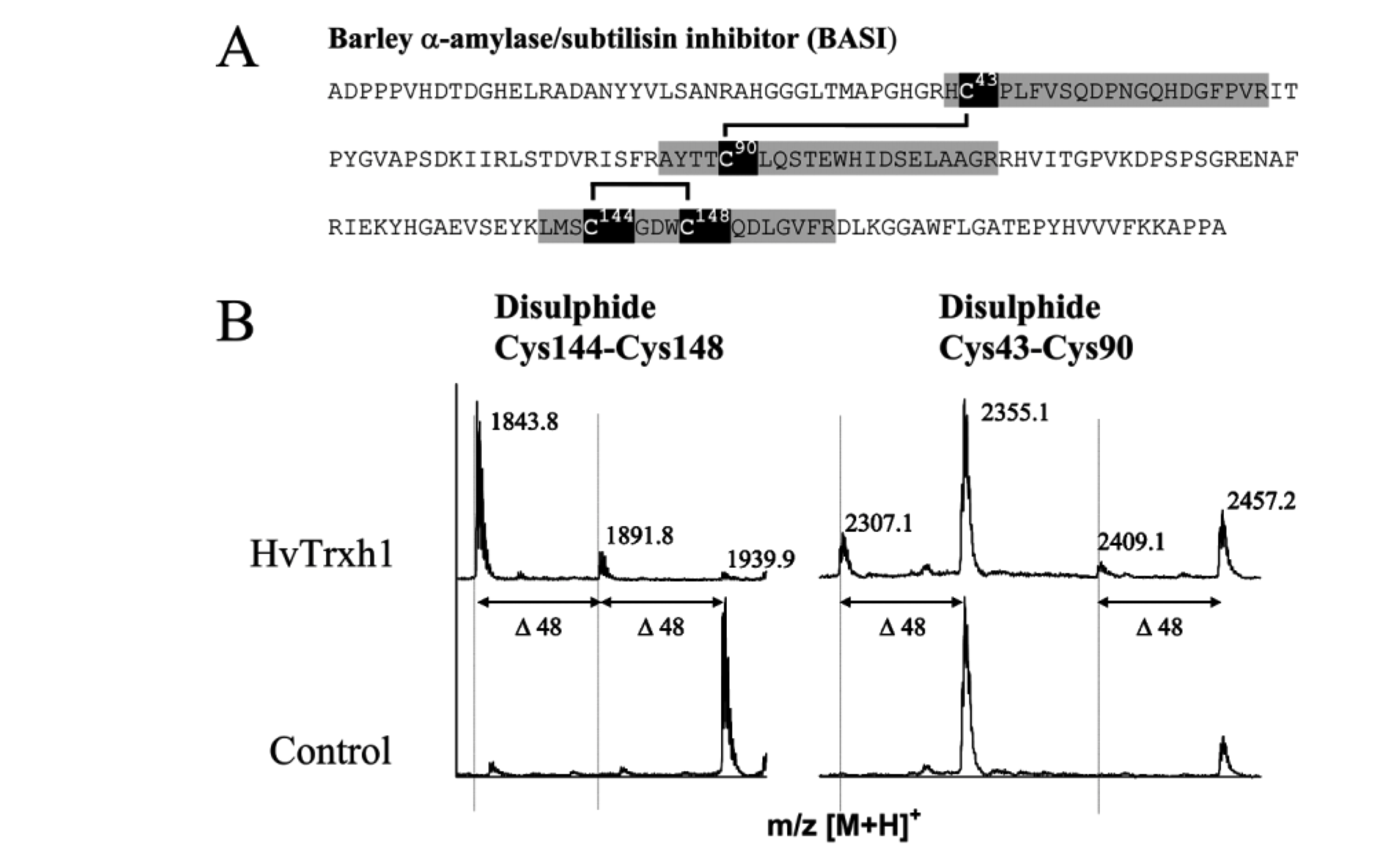
Differential Oxidation (Maeda 2004)
- Target protein: BASI (barley α-amylase/subtilisin inhibitor)
- Thioredoxin h treatment (NADPH + NTR + Trx-h)
- Alkylation using IAA (iodoacetamide; carbamidomethylation (57 Da))
- SDS-PAGE; excision of pieces containing BASI
- Incubation with DTT (reduction of non-target disulphide bonds)
- Alkylation using 4-VP (4-vinylpyridine; pyridylethylation (105 Da))
Differential Oxidation (Maeda 2004)
- After treatment with Trx-h, the accessible cysteinyl side chains in thiol form are carbamidomethylated by IAA (57 Da) whereas those inaccessible or forming disulphides are pyridylethylated by 4-VP (105 Da)
- Control: no Trx-h treatment; cysteines that should be carbamidomethylated (57 Da) are now pyridylethylated (105 Da)
- Compare the MS maps (with & without Trx-h reduction); look for 48 Da differences (105 - 57 = 48)
References
Geigenberger, Peter, Ina Thormählen, Danilo M. Daloso, and Alisdair R. Fernie. 2017. “The Unprecedented Versatility of the Plant Thioredoxin System.” Trends in Plant Science 22 (3): 249–62. https://doi.org/https://doi.org/10.1016/j.tplants.2016.12.008.
Hägglund, Per, Christine Finnie, Hiroyuki Yano, Azar Shahpiri, Bob B. Buchanan, Anette Henriksen, and Birte Svensson. 2016. “Seed Thioredoxin H.” Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 1864 (8): 974–82. https://doi.org/https://doi.org/10.1016/j.bbapap.2016.02.014.
Kobrehel, K, J H Wong, A Balogh, F Kiss, B C Yee, and B B Buchanan. 1992. “Specific Reduction of Wheat Storage Proteins by Thioredoxin H.” Plant Physiology 99 (3). American Society of Plant Biologists: 919–24. https://doi.org/10.1104/pp.99.3.919.
Maeda, Kenji, Christine Finnie, and Birte Svensson. 2005. “Identification of Thioredoxin h-Reducible Disulphides in Proteomes by Differential Labelling of Cysteines: Insight into Recognition and Regulation of Proteins in Barley Seeds by Thioredoxin h.” PROTEOMICS 5 (6): 1634–44. https://doi.org/10.1002/pmic.200401050.
Motohashi, Ken, Aiko Kondoh, Michael T. Stumpp, and Toru Hisabori. 2001. “Comprehensive Survey of Proteins Targeted by Chloroplast Thioredoxin.” Proceedings of the National Academy of Sciences 98 (20). National Academy of Sciences: 11224–9. https://doi.org/10.1073/pnas.191282098.
Verdoucq, L, F Vignols, J P Jacquot, Y Chartier, and Y Meyer. n.d. “In Vivo Characterization of a Thioredoxin H Target Protein Defines a New Peroxiredoxin Family.” J Biol Chem 274 (28). Laboratoire de Physiologie et de Biologie Moléculaire des Plantes, UMR 5545, Université de Perpignan, Avenue de Villeneuve, F 66025, Perpignan, France.: 19714–22. https://doi.org/10.1074/jbc.274.28.19714.
Yamazaki, Daisuke, Ken Motohashi, Takeshi Kasama, Yukichi Hara, and Toru Hisabori. n.d. “Target Proteins of the Cytosolic Thioredoxins in Arabidopsis Thaliana.” Plant Cell Physiol 45 (1). Chemical Resources Laboratory, Tokyo Institute of Technology, Nagatsuta 4259, Midori-ku, Yokohama, 226-8503 Japan.: 18–27. https://doi.org/10.1093/pcp/pch019.